# import os
# import numpy as np
import pandas as pd
# import seaborn as sns
# import matplotlib.pyplot as plt
from skimpy import skim
import plotly.express as pxHousing Regression
This dataset is composed of over 20,000 rows and 9 columns.
Notebook goals:
- Complete regression analysis with train-test split and two models for comparison
- Data investigation of summary statistics and visualizations
- Metric evaluation and performance visualization
Imports
from sklearn.datasets import fetch_california_housing
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
from sklearn.ensemble import RandomForestRegressor
from sklearn.metrics import (
mean_squared_error,
explained_variance_score
)Data
Get the Data
- Import the data from sklearn
- Transfer data into pandas DataFrame
- Basic data overview
Note: Data is returned as a bunch object, similar to a dictionary. We’ll convert it to a pandas df.
# Load data
data = fetch_california_housing()type(data)sklearn.utils._bunch.Bunchdata.keys()dict_keys(['data', 'target', 'frame', 'target_names', 'feature_names', 'DESCR'])# Convert data to pandas dataframe
df = pd.DataFrame(data=data['data'], columns=data['feature_names'])df.head()| MedInc | HouseAge | AveRooms | AveBedrms | Population | AveOccup | Latitude | Longitude | |
|---|---|---|---|---|---|---|---|---|
| 0 | 8.3252 | 41.0 | 6.984127 | 1.023810 | 322.0 | 2.555556 | 37.88 | -122.23 |
| 1 | 8.3014 | 21.0 | 6.238137 | 0.971880 | 2401.0 | 2.109842 | 37.86 | -122.22 |
| 2 | 7.2574 | 52.0 | 8.288136 | 1.073446 | 496.0 | 2.802260 | 37.85 | -122.24 |
| 3 | 5.6431 | 52.0 | 5.817352 | 1.073059 | 558.0 | 2.547945 | 37.85 | -122.25 |
| 4 | 3.8462 | 52.0 | 6.281853 | 1.081081 | 565.0 | 2.181467 | 37.85 | -122.25 |
df.shape(20640, 8)Add target to df
df["target"] = data['target']df.head()| MedInc | HouseAge | AveRooms | AveBedrms | Population | AveOccup | Latitude | Longitude | target | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 8.3252 | 41.0 | 6.984127 | 1.023810 | 322.0 | 2.555556 | 37.88 | -122.23 | 4.526 |
| 1 | 8.3014 | 21.0 | 6.238137 | 0.971880 | 2401.0 | 2.109842 | 37.86 | -122.22 | 3.585 |
| 2 | 7.2574 | 52.0 | 8.288136 | 1.073446 | 496.0 | 2.802260 | 37.85 | -122.24 | 3.521 |
| 3 | 5.6431 | 52.0 | 5.817352 | 1.073059 | 558.0 | 2.547945 | 37.85 | -122.25 | 3.413 |
| 4 | 3.8462 | 52.0 | 6.281853 | 1.081081 | 565.0 | 2.181467 | 37.85 | -122.25 | 3.422 |
# NOTE: 1 additional column
df.shape(20640, 9)Explore Dataset
df.info()<class 'pandas.core.frame.DataFrame'>
RangeIndex: 20640 entries, 0 to 20639
Data columns (total 9 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 MedInc 20640 non-null float64
1 HouseAge 20640 non-null float64
2 AveRooms 20640 non-null float64
3 AveBedrms 20640 non-null float64
4 Population 20640 non-null float64
5 AveOccup 20640 non-null float64
6 Latitude 20640 non-null float64
7 Longitude 20640 non-null float64
8 target 20640 non-null float64
dtypes: float64(9)
memory usage: 1.4 MBskim(df)╭──────────────────────────────────────────────── skimpy summary ─────────────────────────────────────────────────╮ │ Data Summary Data Types │ │ ┏━━━━━━━━━━━━━━━━━━━┳━━━━━━━━┓ ┏━━━━━━━━━━━━━┳━━━━━━━┓ │ │ ┃ Dataframe ┃ Values ┃ ┃ Column Type ┃ Count ┃ │ │ ┡━━━━━━━━━━━━━━━━━━━╇━━━━━━━━┩ ┡━━━━━━━━━━━━━╇━━━━━━━┩ │ │ │ Number of rows │ 20640 │ │ float64 │ 9 │ │ │ │ Number of columns │ 9 │ └─────────────┴───────┘ │ │ └───────────────────┴────────┘ │ │ number │ │ ┏━━━━━━━━━━━━━━┳━━━━━┳━━━━━━━┳━━━━━━━━━━┳━━━━━━━━━━┳━━━━━━━━━┳━━━━━━━━━┳━━━━━━━━━┳━━━━━━━━┳━━━━━━━━━┳━━━━━━━━┓ │ │ ┃ column ┃ NA ┃ NA % ┃ mean ┃ sd ┃ p0 ┃ p25 ┃ p50 ┃ p75 ┃ p100 ┃ hist ┃ │ │ ┡━━━━━━━━━━━━━━╇━━━━━╇━━━━━━━╇━━━━━━━━━━╇━━━━━━━━━━╇━━━━━━━━━╇━━━━━━━━━╇━━━━━━━━━╇━━━━━━━━╇━━━━━━━━━╇━━━━━━━━┩ │ │ │ MedInc │ 0 │ 0 │ 3.871 │ 1.9 │ 0.4999 │ 2.563 │ 3.535 │ 4.743 │ 15 │ ▆█▂ │ │ │ │ HouseAge │ 0 │ 0 │ 28.64 │ 12.59 │ 1 │ 18 │ 29 │ 37 │ 52 │ ▂▆███▅ │ │ │ │ AveRooms │ 0 │ 0 │ 5.429 │ 2.474 │ 0.8462 │ 4.441 │ 5.229 │ 6.052 │ 141.9 │ █ │ │ │ │ AveBedrms │ 0 │ 0 │ 1.097 │ 0.4739 │ 0.3333 │ 1.006 │ 1.049 │ 1.1 │ 34.07 │ █ │ │ │ │ Population │ 0 │ 0 │ 1425 │ 1132 │ 3 │ 787 │ 1166 │ 1725 │ 35680 │ █ │ │ │ │ AveOccup │ 0 │ 0 │ 3.071 │ 10.39 │ 0.6923 │ 2.43 │ 2.818 │ 3.282 │ 1243 │ █ │ │ │ │ Latitude │ 0 │ 0 │ 35.63 │ 2.136 │ 32.54 │ 33.93 │ 34.26 │ 37.71 │ 41.95 │ █▃▁▆▁ │ │ │ │ Longitude │ 0 │ 0 │ -119.6 │ 2.004 │ -124.3 │ -121.8 │ -118.5 │ -118 │ -114.3 │ ▁▆▂█▃ │ │ │ │ target │ 0 │ 0 │ 2.069 │ 1.154 │ 0.15 │ 1.196 │ 1.797 │ 2.647 │ 5 │ ▄█▆▃▂▂ │ │ │ └──────────────┴─────┴───────┴──────────┴──────────┴─────────┴─────────┴─────────┴────────┴─────────┴────────┘ │ ╰────────────────────────────────────────────────────── End ──────────────────────────────────────────────────────╯
Train-Test Split
Splitting data before EDA can be helpful to avoid data leakage or incorrect assumptions about what the data shows.
EDA and training will use only the train data. Test data will be used for evaluation only.
Note: Splitting is normally done with X (features) and y (target) separated to avoid data leakage. Here, we will training and test data and split X, y before model training.
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33, random_state=42)
train_df, test_df = train_test_split(df, test_size=0.33, random_state=42)EDA
print(f'Shape of original data: {df.shape}')
print(f'Shape of training data: {train_df.shape}')
print(f'Shape of training data: {test_df.shape}')Shape of original data: (20640, 9)
Shape of training data: (13828, 9)
Shape of training data: (6812, 9)print(f'Percent of data in training: {len(train_df)/len(df):.0%}')
print(f'Percent of data in test: {len(test_df)/len(df):.0%}')Percent of data in training: 67%
Percent of data in test: 33%Focus on Train for EDA
It is best not to peek at test data. It can lead to unsupported assumptions.
skim(train_df)╭──────────────────────────────────────────────── skimpy summary ─────────────────────────────────────────────────╮ │ Data Summary Data Types │ │ ┏━━━━━━━━━━━━━━━━━━━┳━━━━━━━━┓ ┏━━━━━━━━━━━━━┳━━━━━━━┓ │ │ ┃ Dataframe ┃ Values ┃ ┃ Column Type ┃ Count ┃ │ │ ┡━━━━━━━━━━━━━━━━━━━╇━━━━━━━━┩ ┡━━━━━━━━━━━━━╇━━━━━━━┩ │ │ │ Number of rows │ 13828 │ │ float64 │ 9 │ │ │ │ Number of columns │ 9 │ └─────────────┴───────┘ │ │ └───────────────────┴────────┘ │ │ number │ │ ┏━━━━━━━━━━━━━━┳━━━━━┳━━━━━━━┳━━━━━━━━━━┳━━━━━━━━━━┳━━━━━━━━━┳━━━━━━━━━┳━━━━━━━━━┳━━━━━━━━┳━━━━━━━━━┳━━━━━━━━┓ │ │ ┃ column ┃ NA ┃ NA % ┃ mean ┃ sd ┃ p0 ┃ p25 ┃ p50 ┃ p75 ┃ p100 ┃ hist ┃ │ │ ┡━━━━━━━━━━━━━━╇━━━━━╇━━━━━━━╇━━━━━━━━━━╇━━━━━━━━━━╇━━━━━━━━━╇━━━━━━━━━╇━━━━━━━━━╇━━━━━━━━╇━━━━━━━━━╇━━━━━━━━┩ │ │ │ MedInc │ 0 │ 0 │ 3.877 │ 1.903 │ 0.4999 │ 2.569 │ 3.539 │ 4.757 │ 15 │ ▅█▂ │ │ │ │ HouseAge │ 0 │ 0 │ 28.56 │ 12.6 │ 1 │ 18 │ 29 │ 37 │ 52 │ ▂▆███▅ │ │ │ │ AveRooms │ 0 │ 0 │ 5.437 │ 2.449 │ 0.8889 │ 4.46 │ 5.232 │ 6.059 │ 141.9 │ █ │ │ │ │ AveBedrms │ 0 │ 0 │ 1.098 │ 0.4457 │ 0.3333 │ 1.007 │ 1.05 │ 1.1 │ 25.64 │ █ │ │ │ │ Population │ 0 │ 0 │ 1431 │ 1146 │ 3 │ 793 │ 1170 │ 1729 │ 35680 │ █ │ │ │ │ AveOccup │ 0 │ 0 │ 3.129 │ 12.65 │ 0.6923 │ 2.432 │ 2.82 │ 3.282 │ 1243 │ █ │ │ │ │ Latitude │ 0 │ 0 │ 35.65 │ 2.134 │ 32.55 │ 33.94 │ 34.27 │ 37.72 │ 41.95 │ █▃▁▆▁ │ │ │ │ Longitude │ 0 │ 0 │ -119.6 │ 2.005 │ -124.3 │ -121.8 │ -118.5 │ -118 │ -114.3 │ ▁▆▂█▃ │ │ │ │ target │ 0 │ 0 │ 2.067 │ 1.154 │ 0.15 │ 1.194 │ 1.792 │ 2.64 │ 5 │ ▄█▆▃▂▂ │ │ │ └──────────────┴─────┴───────┴──────────┴──────────┴─────────┴─────────┴─────────┴────────┴─────────┴────────┘ │ ╰────────────────────────────────────────────────────── End ──────────────────────────────────────────────────────╯
All Features and Target are Numeric
A loop can help iterate over the columns and produce visualizations.
col_list = train_df.columns.to_list()col_list['MedInc',
'HouseAge',
'AveRooms',
'AveBedrms',
'Population',
'AveOccup',
'Latitude',
'Longitude',
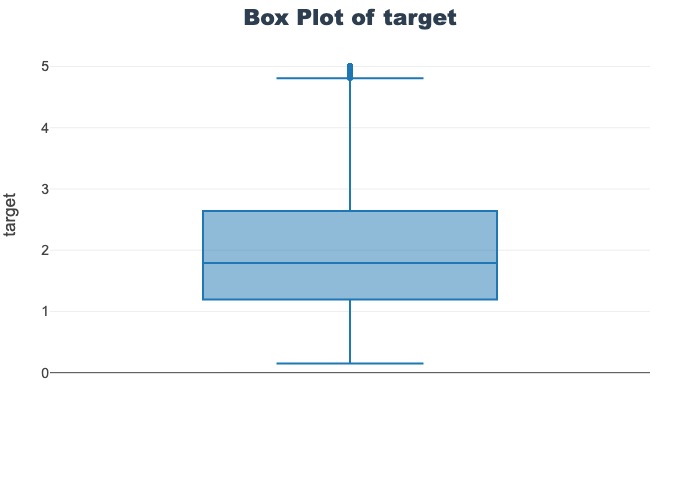
'target']for col in col_list:
fig = px.box(train_df, y=col, title=f'Box Plot of {col}')
fig.show()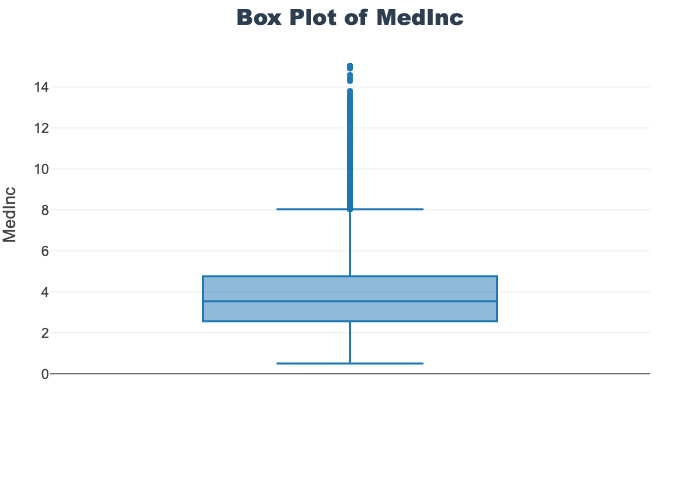
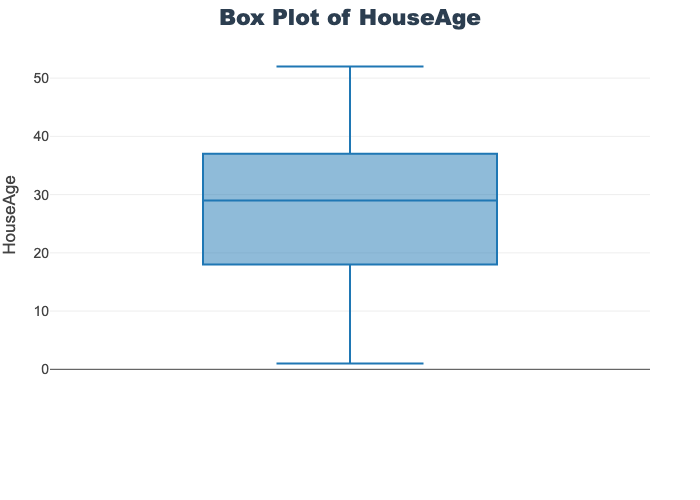
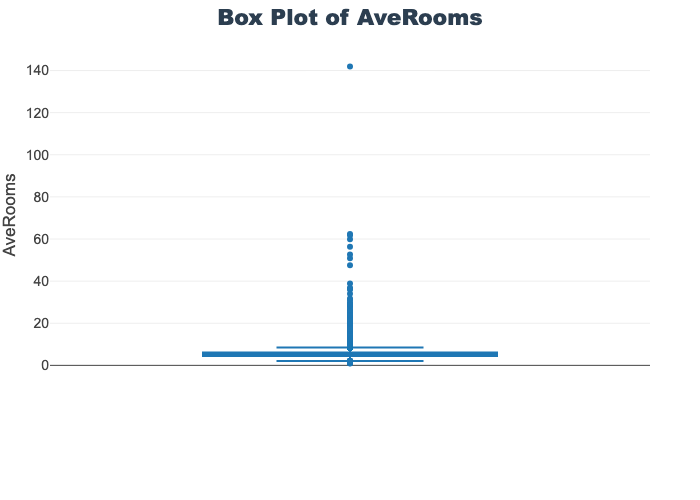
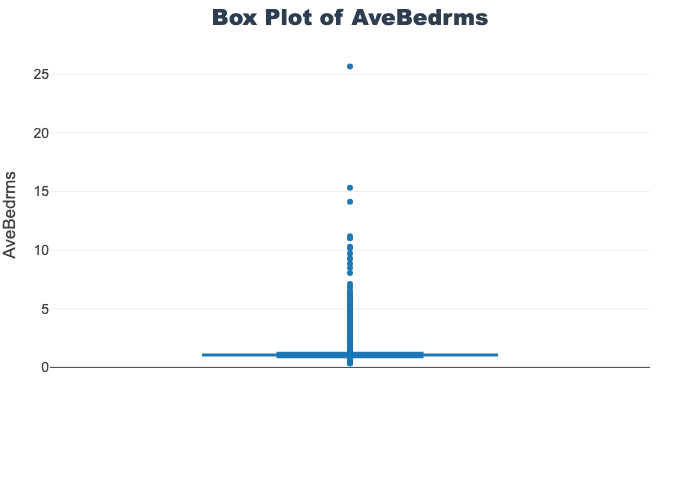
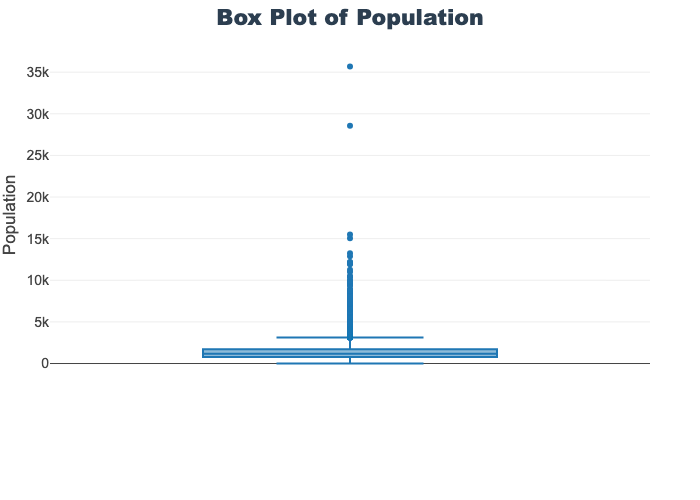
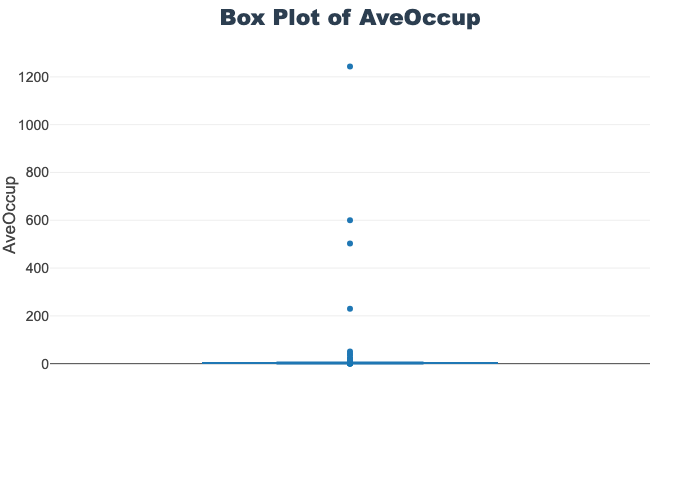
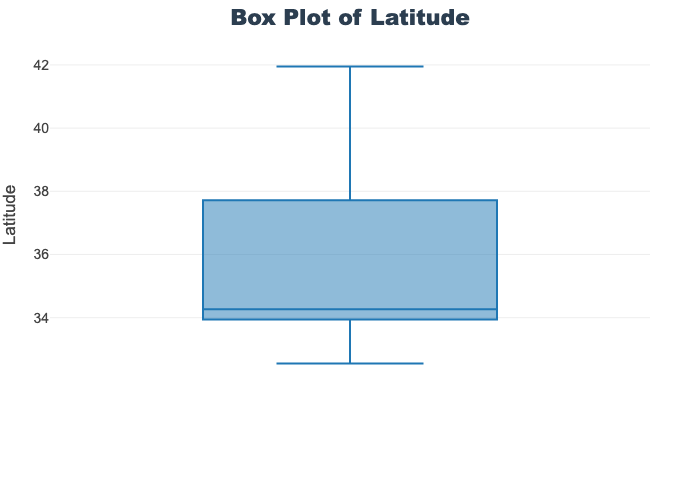
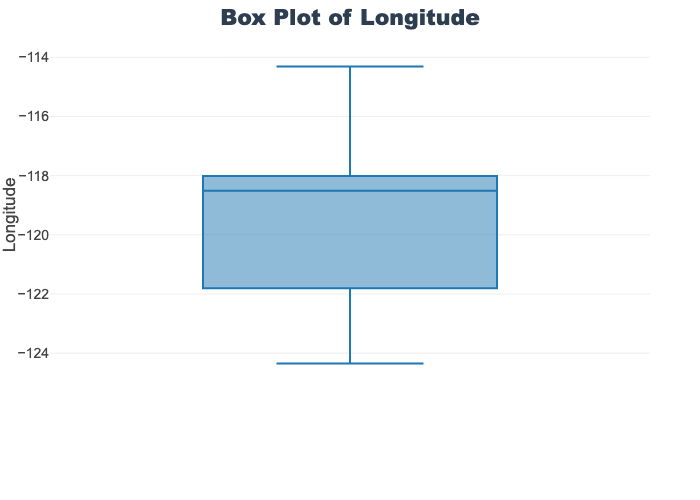
# px.box(sepal_df, x="species", y="value",
# facet_col="variable", title="Sepal Features by Species",
# color="species")train_df.columnsIndex(['MedInc', 'HouseAge', 'AveRooms', 'AveBedrms', 'Population', 'AveOccup',
'Latitude', 'Longitude', 'target'],
dtype='object')Model Training
# Instantiate model, fit model, save model
lr = LinearRegression()
lr.fit(X_train, y_train)--------------------------------------------------------------------------- NameError Traceback (most recent call last) Cell In[21], line 3 1 # Instantiate model, fit model, save model 2 lr = LinearRegression() ----> 3 lr.fit(X_train, y_train) NameError: name 'X_train' is not defined
# Instantiate model, fit model, save model
rf = RandomForestRegressor()
rf.fit(X_train, y_train)# Predict on train with both models
# NOTE: test metrics are more insightful
lr_train_preds = lr.predict(X_train)
rf_train_preds = rf.predict(X_train)# Calculate mean squared error for both models
lr_mse = mean_squared_error(y_train, lr_train_preds)
rf_mse = mean_squared_error(y_train, rf_train_preds)# Print calculations
print(f"The MSE for the linear regression models is : {lr_mse: .2f}")
print(f"The MSE for the random forest regression models is : {rf_mse: .2f}")# Plot both predictions
plt.figure(figsize=(10,10))
plt.scatter(y_train, lr_train_preds, c='crimson', label='Linear Regression')
plt.scatter(y_train, rf_train_preds, c='gold', label='RF Regression')
plt.xlabel('True Values', fontsize=15)
plt.ylabel('Predictions', fontsize=15)
plt.title('Training Error', fontsize=15)
plt.legend()
plt.tight_layout()
plt.show()Evaluate Models Notebook
# linear regression predict
lr_preds = lr.predict(X_test)
lr_preds# random forest regression predict
rf_preds = rf.predict(X_test)
rf_preds# Calculate explained variance for both models
lr_evs = explained_variance_score(y_test, lr_preds)
rf_evs = explained_variance_score(y_test, rf_preds)# Display explained variance scores
print(f'The explained variance score for the linear regression models is: {lr_evs: .2f}')
print(f'The explained variance score for the random forest regression models is: {rf_evs: .2f}')# Calculate mean squared error (MSE)
lr_mse = mean_squared_error(y_test, lr_preds)
rf_mse = mean_squared_error(y_test, rf_preds)# Display MSE
print(f"The MSE for the linear regression models is : {lr_mse: .2f}")
print(f"The MSE for the random forest regression models is : {rf_mse: .2f}")# create y_df with real and predicted values
y_df=pd.DataFrame({'y_true': y_test, 'lr_preds': lr_preds, 'rf_preds': rf_preds})# Check df
y_df.head()# Get correlation across real, lr, and rf values
y_df.corr()# Seaborn pair plot on y data
sns.pairplot(y_df)# Plot results
plt.figure(figsize=(10,10))
plt.scatter(y_test.target, lr_preds, c='crimson', label='Linear Regression')
plt.scatter(y_test.target, rf_preds, c='gold', label='RF Regression')
plt.xlabel('True Values', fontsize=15)
plt.ylabel('Predictions', fontsize=15)
plt.title('Test Error', fontsize=15)
plt.legend()
plt.tight_layout()
plt.show()